Sustainable Production and Characterization of a Novel β-1,3-1,4-Glucanase from Aspergillus niger CCUG33991 for Enhanced Animal Feed and Nutraceutical Applications
Keywords:
Animal feed Industry, Aspergillus niger, beta-1,3-1,4-Glucanase, Cereal Structure Modification Solid-State Fermentation, Sustainable Biotechnology, Wheat BranAbstract
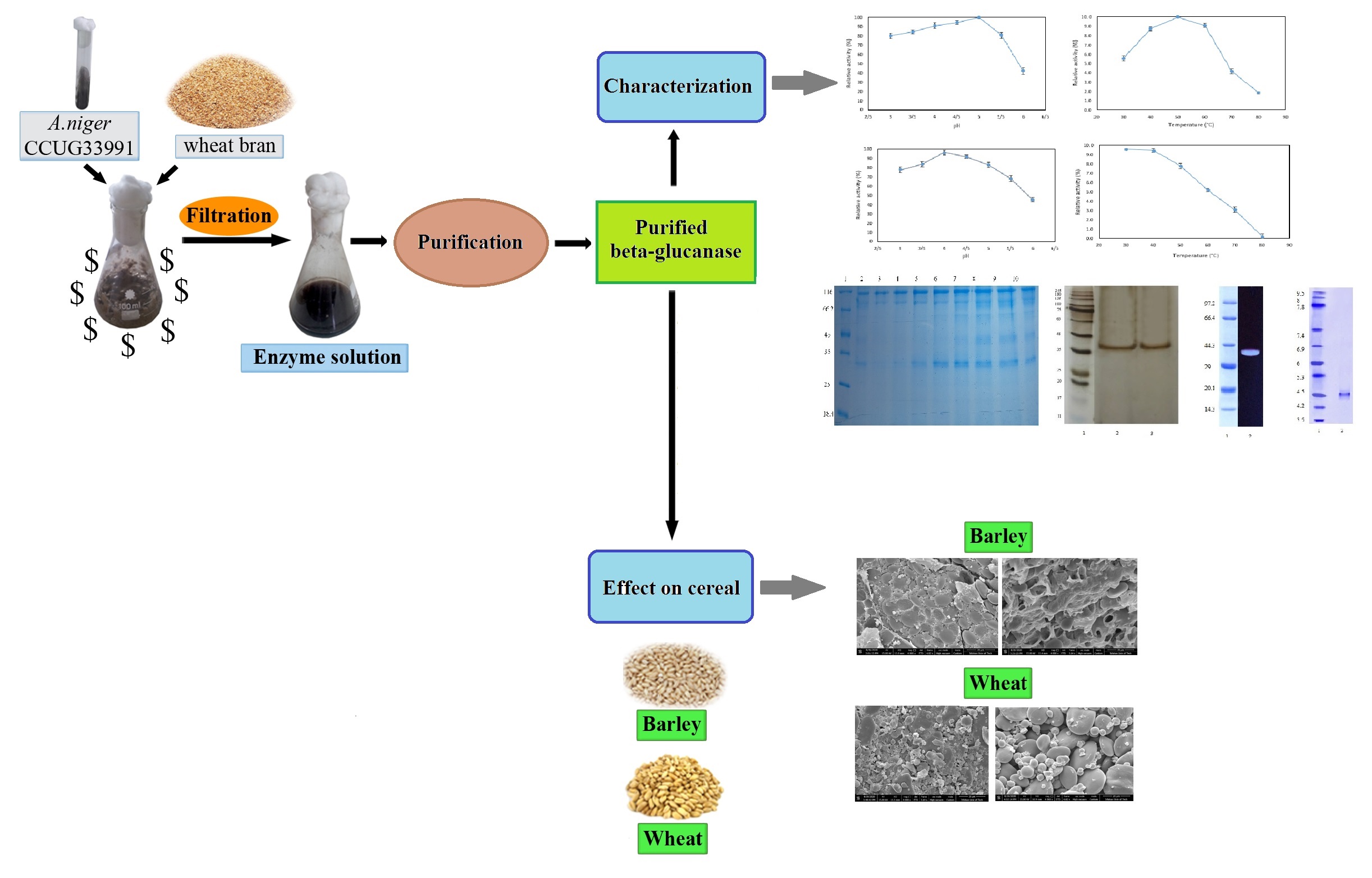
With global demands for sustainable food systems intensifying, cost-effective enzyme production offers transformative solutions for the animal feed industry. β-1,3-1,4-Glucanase from Aspergillus niger CCUG33991 was purified 12.3-fold with a 20.3% yield using ammonium sulfate precipitation and size exclusion chromatography. This 39 kDa enzyme, with an isoelectric point of 4.5, exhibits optimal activity at pH 5.0 and 50°C, with robust stability at low pH and temperatures below 40°C. Kinetic studies revealed a Km of 5.1 mg/ml and Vm of 52.1 µmol/min/mg. The enzyme’s sensitivity to Mn²⁺ ions and its ability to modify cereal structures enhance its potential to improve feed digestibility and address sanitary issues in poultry. Utilizing wheat bran in solid-state fermentation, this study presents a sustainable, scalable approach with applications in animal nutrition, brewing, and emerging fields like microbiome engineering and nutraceuticals.
Downloads
References
1. Tang Y, Yang S, Yan Q, Zhou P, Cui J, Jiang Z. Purification and characterization of a novel β-1,3;1,4-glucanase (Lichenase) from thermophilic Rhizomucor miehei with high specific activity and its gene sequence. J Agric Food Chem. 2012;60:2354-61.
2. Yang S, Qiaojuan Y, Jiang Z, Fan G, Wang L. Biochemical characterization of a novel thermostable beta-1,3-1,4-glucanase (lichenase) from Paecilomyces thermophila. J Agric Food Chem. 2008;56:5345-51.
3. Celestino KRS, Cunha RB, Felix CR. Characterization of a β-glucanase produced by Rhizopus microsporus var. microsporus, and its potential for application in the brewing industry. BMC Biochem. 2006;7:1-9.
4. Beckmann L, Simon O, Vahjen W. Isolation and identification of mixed linked β-glucan degrading bacteria in the intestine of broiler chickens and partial characterization of respective 1,3-1,4-β-glucanase activities. J Basic Microbiol. 2006;46:175-85.
5. Bhat MK, Bhat S. Cellulose degrading enzymes and their potential industrial applications. Biotechnol Adv. 1997;15:583-620.
6. Kirk O, Borchert TV, Fuglsang CC. Industrial enzyme applications. Curr Opin Biotechnol. 2002;13:345-51.
7. Anish R, Rahman MS, Rao M. Application of cellulases from an alkalothermophilic Thermomonospora sp. in biopolishing of denims. Biotechnol Bioeng. 2007;96:48-56.
8. Kotchoni SO, Shonukan OO. Regulatory mutations affecting the synthesis of cellulase in Bacillus pumilus. World J Microbiol Biotechnol. 2002;18:487-91.
9. Bai H, Wang H, Sun J, Irfan M, Han M, Huang Y, et al. Purification and characterization of beta 1,4-glucanases from Penicillium simplicissimum H-11. BioRes. 2013;8:3657-71.
10. Singhania RR, Sukumaran RK, Patel AK, Larroche C, Pandey A. Advancement and comparative profiles in the production technologies using solid-state and submerged fermentation for microbial cellulases. Enzyme Microb Technol. 2010;46:541-9.
11. Gao J, Weng H, Zhu D, Yuan M, Guan F, Xi Y. Production and characterization of cellulolytic enzymes from the thermoacidophilic fungal Aspergillus terreus M11 under solid-state cultivation of corn stover. Bioresour Technol. 2008;99:7623-9.
12. Soma M, Rangasamy M. Production of cellulase by Aspergillus niger under submerged and solid state fermentation using coir waste as a substrate. Braz J Microbiol. 2011;42:1119-27.
13. Prajanban J, Thongkhib C, Kitpreechavanich V. Selection of high β-glucanase produced Aspergillus strain and factors affecting the enzyme production in solid-state fermentation. Kasetsart J. 2008;42:294-9.
14. Jian-yi S, Wei-fen L, Zi-rong X, Sai-hong G. Purification and some properties of a β-glucanase from a strain, Trichoderma reesei GXC. J Zheijang Univ-Sci. 2002;3:106-12.
15. Shahryari Z, Fazaelipoor MH, Setoode HP, Nair RB, Taherzadeh MJ, Ghasemi Y. Utilization of wheat straw for fungal phytase production. Int J Recycl Org Waste Agric. 2018;7:345-55.
16. Chaari F, Bhiri F, Blibech M, Maktouf S, Ellouz-Chaabouni S, Ellouz-Ghorbel R. Potential application of two thermostable lichenases from a newly isolated Bacillus licheniformis UEB CF: purification and characterization. Process Biochem. 2012;47:509-16.
17. Maktouf S, Moulis C, Kamoun A, Chaari F, Chaabouni SE, Remaud-Simeon M. A laundry detergent compatible lichenase: statistical optimization for production under solid state fermentation on crude millet. Ind Crops Prod. 2013;43:349-54.
18. Chaari F, Kamoun A, Bhiri F, Blibech M, Ellouze-Ghorbel R, Ellouz-Chaabouni S. Statistical optimization for the production of lichenase by a newly isolated Bacillus licheniformis UEB CF in solid state fermentation using pea pomace as a novel solid support. Ind Crops Prod. 2012;40:192-8.
19. Yang S, Xiong H, Yan Q, Yang H, Jiang Z. Purification and characterization of a novel alkaline β-1,3-1,4-glucanase (lichenase) from thermophilic fungus Malbranchea cinnamomea. J Ind Microbiol Biotechnol. 2014;41:1487-95.
20. Elgharbi F, Hmida-Sayari A, Sahnoun M, Kammoun R, Jlaeil L, Hassairi H, et al. Purification and biochemical characterization of a novel thermostable lichenase from Aspergillus niger US368. Carbohydr Polym. 2013;98:967-75.
21. Chaari F, Belghith-Fendri L, Blibech M, Driss D, Ellouzia SZ, Sameh M, et al. Biochemical characterization of a lichenase from Penicillium occitanis Pol6 and its potential application in the brewing industry. Process Biochem. 2014;49:1040-6.
22. Heidary-Vinche M, Khanahmadi M, Ataei SA, Danafar F. Optimization of process variables for production of beta-glucanase by Aspergillus niger CCUG33991 in solid-state fermentation using wheat bran. Waste Biomass Valorization. 2020;11.
23. Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426-8.
24. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265-75.
25. Lloberas J, Querol E, Bernués J. Purification and characterization of endo-β-1,3-1,4-d-glucanase activity from Bacillus licheniformis. Appl Microbiol Biotechnol. 1988;29:32-8.
26. Laemmli UK. Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature. 1970;227:680-5.
27. Tervila-Wilo A, Parkkonen T, Morgan A, Hopeakoski-Nurminen M, Poutanen K, Heikkinen P, et al. In vitro digestion of wheat microstructure with xylanase and cellulase from Trichoderma reesei. J Cereal Sci. 1996;24:215-25.
28. Planas A. Bacterial 1,3-1,4-beta-glucanases: structure, function and protein engineering. Biochim Biophys Acta. 2000;1543:361-82.
29. Hong J, Tamaki H, Yamamoto K, Kumagai H. Cloning of a gene encoding a thermo-stable endo-β-1,4-glucanase from Thermoascus aurantiacus and its expression in yeast. Biotechnol Lett. 2003;25:657-61.
30. Anish R, Rao M. Biochemical characterization of a novel β-1,3-1,4 glucan 4-glucanohydrolase from Thermomonospora sp. having a single active site for lichenan and xylan. Biochimie. 2007;89:1489-97.
31. Murray PG, Grassick A, Laffey CD, Cuffe MM, Higgins T, Savage AV, et al. Isolation and characterization of a thermostable endo-beta-glucanase active on 1,3-1,4-beta-D-glucans from the aerobic fungus talaromyces emersonii CBS 814.70. Enzyme Microb Technol. 2001;29:90-8.
32. Boyce A, Walsh AG. Production, purification and application-relevant characterisation of an endo-1,3(4)-β-glucanase from Rhizomucor miehei. Appl Microbiol Biotechnol. 2007;76:835-41.
33. Erfle JD, Teather RM, Wood PJ, Irvin JE. Purification and properties of a 1,3-1,4-beta-D-glucanase (lichenase, 1,3-1,4-beta-D-glucan 4-glucanohydrolase, EC 3.2.1.73) from Bacteroides succinogenes cloned in Escherichia coli. The Biochemical Journal. 1988;255:833-41.
34. Shah AR, Madamwar D. Xylanase production under solid-state fermentation and its characterization by an isolated strain of Aspergillus foetidus in India. World J Microbiol Biotechnol. 2005;21:233-43.
35. Ezio AM, Edward AH, Edward LR. Enzymatic Properties of a B-glucanase from Bacillus subtilis. J Biol Chem. 1961;236:2858-62.
36. Spilliaert R, Hreggvidsson GO, Kristjansson JK, Eggertsson G, Palsdottir A. Cloning and sequencing of a Rhodothermus marinus gene, bglA, coding for a thermostable beta-glucanase and its expression in Escherichia coli. Eur J Biochem. 1994;224:923-30.
37. Sharma A, Nakas JP. Preliminary characterization of laminarinase from Trichoderma longibrachiatum. Enzyme Microb Technol. 1987;9:89-93.
38. McCleary BV, Nurthen E. Measurement of (1 → 3)(1 → 4)- β -D-glucan in malt, wort and beer. J Inst Brew. 1986;92:168-73.
39. loprete D, Hill T. Isolation and Characterization of an Endo-(1,4)-β-Glucanase Secreted by Achlya ambisexualis. Mycologia. 2002;94:903-11.
40. Grishutin SG, Gusakov AV, Dzedzyulya EL, Sinitsyn AP. A lichenase-like family 12 endo-(1→4)-beta-glucanase from Aspergillus japonicus: study of the substrate specificity and mode of action on beta-glucans in comparison with other glycoside hydrolases. Carbohydr Res. 2006;341:218-29.
41. MacRitchie F. Concepts in Cereal Chemistry. Manhattan, USA2010. 1-421 p.
42. Hong MR, Kim YS, Joo AR, Lee JK, Kim YS, Oh DK. Purification and characterization of a thermostable beta-1,3-1,4-glucanase from Laetiporus sulphureus var. miniatus. J Microbiol Biotechnol. 2009;19:818-22.
43. Chen H, Li XL, Ljungdahl LG. Sequencing of a 1,3-1,4-beta-D-glucanase (lichenase) from the anaerobic fungus Orpinomyces strain PC-2: properties of the enzyme expressed in Escherichia coli and evidence that the gene has a bacterial origin. J Bacteriol. 1977;179:6028-34.
44. Ahmad A, Anjum FM, Zahoor T, Nawaz H, Ahmed Z. Extraction and characterization of β-glucan from oat for industrial utilization. Int J Biol Macromol. 2010;46:304-9.
45. McCarthy T, Hanniffy Q, Savage AV, Tuohy MG. Catalytic properties and mode of action of three endo-beta-glucanases from Talaromyces emersonii on soluble beta-1,4- and beta-1,3,1,4-linked glucans. Int J Biol Macromol. 2003;33:141-8.
46. Demirbas A. β-Glucan and mineral nutrient contents of cereals grown in Turkey. Food Chem. 2005;90:773-7.
47. Holopainen-Mantila U, Raulio M. Cereal Grain Structure by Microscopic Analysis. Imaging Technologies and Data Processing for Food Engineers2016. p. 1-39.
48. Teymouri H, Zargh H, Golian A. Evaluation of Hull-Less barley with or without enzyme, cocktail in the finisher diets of broiler chickens. J Agr Sci Tech. 2018;20:469-83.
49. Zhao Z, Ramachandran P, Kim TS, Chen Z, Jeya M, Lee JK. Characterization of an acid-tolerant β-1,4-glucosidase from Fusarium oxysporum and its potential as an animal feed additive. Appl Microbiol Biotechnol. 2013;97:10003-11.
Downloads
Published
Submitted
Revised
Accepted
Issue
Section
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.