Molecular Docking Study of Small Plant-based Molecules for Inhibiting PBP2a Protein in Methicillin-Resistant Staphylococcus aureus (MRSA)
Keywords:
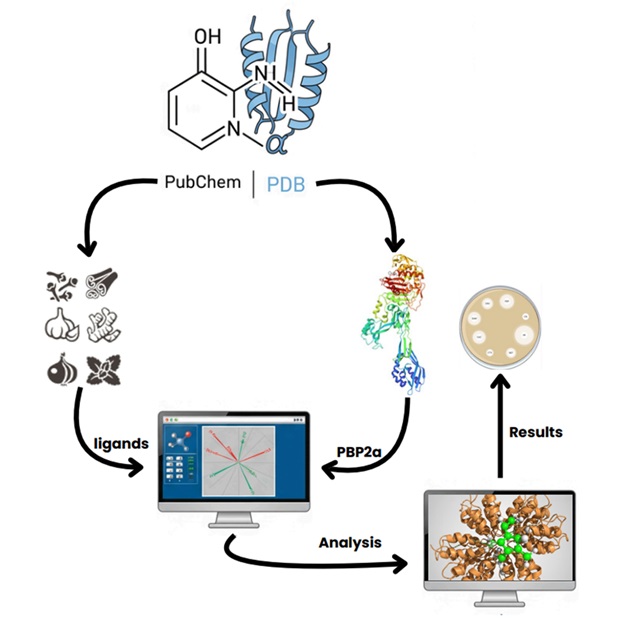
Antibiotic resistance, Methicillin-Resistant Staphylococcus aureus (MRSA), Plant-based compounds, Curcumin, Quercetin, Molecular docking, In silico, Drug discoveryAbstract
Downloads
Download data is not yet available.
Downloads
Published
2025-11-23
Submitted
2024-09-20
Revised
2024-11-19
Accepted
2024-11-22
Issue
Section
Short communication
License
Copyright (c) 2025 Dr. Parvin Shariati; Mr. Reza Nori (Author)

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
How to Cite
Shariati, P., & Nori, R. (2025). Molecular Docking Study of Small Plant-based Molecules for Inhibiting PBP2a Protein in Methicillin-Resistant Staphylococcus aureus (MRSA). BiotechIntellect, 2(1), e16 (1-8). https://doi.org/10.61882/BiotechIntellect.2.1.5