Investigation of Antibiotic Resistance Patterns of Klebsiella pneumoniae Isolates from Educational and Medical Centers of Mazandaran University of Medical Sciences, 2020-21
Keywords:
Klebsiella pneumoniae , Antibiotic Resistance , Mazandaran, IranAbstract
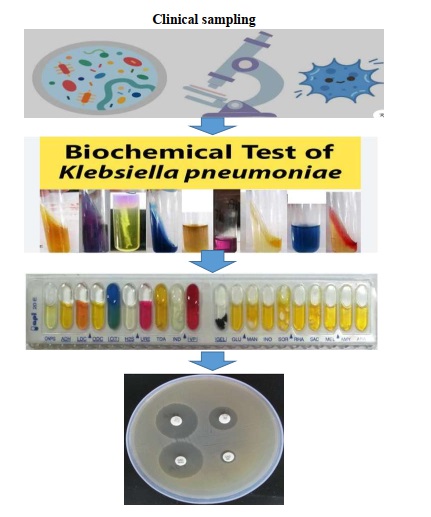
Klebsiella pneumoniae is a Gram-negative, non-motile, encapsulated bacilli, Klebsiella pneumoniae acts as an opportunistic pathogen in the occurrence of hospital-acquired infections. This bacterium is often resistant to several classes of antibiotics, including beta-lactams. The aim of this study was to investigate the frequency and antibiotic resistance of Klebsiella pneumoniae strains isolated from hospitalized patients in the educational and medical centers of Mazandaran University of Medical Sciences from 2020-2021. In this descriptive-cross-sectional study, 50 Klebsiella pneumoniae strains were collected from various clinical samples of hospitalized patients in educational and medical centers affiliated with Mazandaran University of Medical Sciences. Sample cultures were performed on Eosin methylene blue and Blood agar media, and after 24 hours of incubation, Gram staining and oxidase tests were conducted on the grown colonies. Gram-negative and oxidase-negative bacilli were identified, and the presence of Klebsiella pneumoniae was confirmed by culturing bacteria on TSI, SIM, MR-VP, citrate, and urea media. Then, antibiotic susceptibility testing was performed using the disk diffusion method according to CLSI guidelines. The antibiotic disks included ciprofloxacin, imipenem, meropenem, cefepime, ceftazidime, gentamicin, amikacin, and ceftriaxone. In this study, among the 50 isolates examined, 61% were resistant to ciprofloxacin, 60% to imipenem, 49% to meropenem, 56% to cefepime, 58% to ceftazidime, 35% to gentamicin, 13% to amikacin, and 55% to ceftriaxone. Amikacin was identified as the most sensitive and effective antibiotic in this study. The results of the current study indicate an increase in the resistance of Klebsiella pneumoniae isolates to various antibiotics compared to previous studies. Further studies in this area can better guide us in confronting the antibiotic resistance of these infectious bacteria. Therefore, for infection control and to prevent the spread of drug-resistant bacteria, precise management in drug prescription and identification of resistant isolates is essential.
Downloads
References
1. Green VL, Verma A, Owens RJ, Phillips SE, Carr SB. Structure of New Delhi metallo-β-lactamase 1 [NDM-1]. Acta Crystallographica Section F: Structural Biology and Crystallization Communications. 2011;67(10):1160-4.
2. Akhavan TF, Eslami G, Zandi H, Mousavi DM, Zarei M. Prevalence of blaVIM, blaIPM and blaNDM Metallo-Beta-Lactamases Enzymes in Pseudomonas aeruginosa Isolated from Burn Wounds in Shahid Sadoughi Burn Hospital, Yazd, Iran. [Journal Title Not Provided]. 2023.
3. Fernández A, Pereira MJ, Suárez JM, Poza M, Treviño M, Villalón P, et al. Emergence in Spain of a multidrug-resistant Enterobacter cloacae clinical isolate producing SFO-1 extended-spectrum β-lactamase. Journal of Clinical Microbiology. 2011;49(3):822-8.
4. Zarei-Yazdeli M, Eslami G, Zandi H, Mousavi SM, Kosha H, Akhavan F, et al. Relationship between antimicrobial resistance and class I integron in Pseudomonas aeruginosa isolated from clinical specimens in Yazd during 2012-2013. Feyz Journal of Kashan University of Medical Sciences. 2014;18(1).
5. Daikos GL, Kosmidis C, Tassios PT, Petrikkos G, Vasilakopoulou A, Psychogiou M, et al. Enterobacteriaceae bloodstream infections: presence of integrons, risk factors, and outcome. Antimicrobial Agents and Chemotherapy. 2007;51(7):2366-72.
6. Kamatchi C, Magesh H, Sekhar U. Identification of clonal clusters of Klebsiella pneumoniae isolates from Chennai by extended spectrum beta lactamase genotyping and antibiotic resistance phenotyping analysis. [Journal Title Not Provided]. 2023.
7. Gootz TD. The global problem of antibiotic resistance. Critical Reviews in Immunology. 2010;30(1):[Page Numbers Not Provided].
8. Ferreira RL, da Silva B, Rezende GS, Nakamura-Silva R, Pitondo-Silva A, Campanini EB, et al. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-lactamase encoding genes in a Brazilian intensive care unit. Frontiers in Microbiology. 2019;9:3198.
9. Pitout JD, Sanders CC, Sanders Jr WE. Antimicrobial resistance with focus on β-lactam resistance in gram-negative bacilli. The American Journal of Medicine. 1997;103(1):51-9.
10. Roch M, Clair P, Renzoni A, Reverdy ME, Dauwalder O, Bes M, et al. Exposure of Staphylococcus aureus to subinhibitory concentrations of β-lactam antibiotics induces heterogeneous vancomycin-intermediate Staphylococcus aureus. Antimicrobial Agents and Chemotherapy. 2014;58(9):5306-14.
11. Karaiskos I, Giamarellou H. Carbapenem-sparing strategies for ESBL producers: when and how. Antibiotics. 2020;9(2):61.
12. Paterson DL, Ko WC, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, et al. Antibiotic therapy for Klebsiella pneumoniae bacteremia: implications of production of extended-spectrum β-lactamases. Clinical Infectious Diseases. 2004;39(1):31-7.
13. Khanfar HS, Bindayna KM, Senok AC, Botta GA. Extended spectrum beta-lactamases [ESBL] in Escherichia coli and Klebsiella pneumoniae: trends in the hospital and community settings. The Journal of Infection in Developing Countries. 2009;3(4):295-9.
14. Shamsazar A, Yousefi-Avarvand A, Khademi F. Antibiotic Resistance Pattern of Klebsiella pneumoniae from Patients in Qaem Teaching Hospital, Mashhad University of Medical Sciences. Human & Environment. 2020;18(1):83-9.
15. Bulman ZP, Krapp F, Pincus NB, Wenzler E, Murphy KR, Qi C, et al. Genomic features associated with the degree of phenotypic resistance to carbapenems in carbapenem-resistant Klebsiella pneumoniae. mSystems. 2021;6(5):e00194-21.
16. Zhou C, Wu Q, He L, Zhang H, Xu M, Yuan B, et al. Clinical and molecular characteristics of carbapenem-resistant hypervirulent Klebsiella pneumoniae isolates in a tertiary hospital in Shanghai, China. Infection and Drug Resistance. 2021;14:2697.
17. Du J, Li P, Liu H, Lü D, Liang H, Dou Y. Phenotypic and molecular characterization of multidrug resistant Klebsiella pneumoniae isolated from a university teaching hospital, China. PLOS ONE. 2014;9(4):e95181.
18. Heidary M, Nasiri MJ, Dabiri H, Tarashi S. Prevalence of drug-resistant Klebsiella pneumoniae in Iran: a review article. Iranian Journal of Public Health. 2018;47(3):317.
19. Khamesipour F, Tajbakhsh E. Analyzed the genotypic and phenotypic antibiotic resistance patterns of Klebsiella pneumoniae isolated from clinical samples in Iran. Emergence. 2016;12:13.
20. Soltan M, Miremadi SA, Sharify Y. Antimicrobial resistance trends of Klebsiella spp. isolated from patients in Imam Khomeini hospital. [Journal Title Not Provided]. 2020.
21. Moghadas AJ, Kalantari F, Sarfi M, Shahhoseini S, Mirkalantari S. Evaluation of Virulence Factors and Antibiotic Resistance Patterns in Clinical Urine Isolates of Klebsiella pneumoniae in Semnan, Iran. Jundishapur Journal of Microbiology. 2021;11(7):1-6.
22. Seifi K, Kazemian H, Heidari H, Rezagholizadeh F, Saee Y, Shirvani F, et al. Evaluation of biofilm formation among Klebsiella pneumoniae isolates and molecular characterization by ERIC-PCR. Jundishapur Journal of Microbiology. 2016;9(1).
23. Dalir A, Razavi S, Talebi M, Masjedian Jazi F, Zahedi Bialvaei A, Mirshekar M. Antibiotic Susceptibility Pattern and Distribution of Virulence Factors among Klebsiella pneumoniae Isolated from Healthy Volunteers. Iran Journal of Medical Microbiology. 2021;15(6):676-83.
24. Ahanjan M, Naderi F, Solimanii A. Prevalence of Beta-lactamases genes and antibiotic resistance pattern of Klebsiella pneumoniae isolated from teaching hospitals, Sari, Iran, 2014. Journal of Mazandaran University of Medical Sciences. 2017;27(149):79-87.
Downloads
Published
Submitted
Revised
Accepted
Issue
Section
License
Copyright (c) 2024 Vajiheh Hazrati Damavandi, Soheila Ahanjan, Mehrdad Gholami, Shahram Divsalar, Ebrahim Nemati Hoolaee (Author); Mohammad Ahanjan; Mia Ayu Gusti (Author)

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.